

Medical image registrationCNRS - Institut de Mathématiques de Toulouse (Jan. 2012 - .)University of Oxford (Jan. 2011 - Jan. 2012)Imperial College London (Jan. 2009 - Dec. 2010)Context:Since January 2009, I work on the development of new techniques for the registration of 3D medical images. Between January 2009 and December 2010 this work has been done in collaboration with Darryl Holm (Institute for Mathematical Sciences) and Daniel Rueckert (Biomedical Image Analysis Group) at Imperial College London, UK. Between January 2011 and January 2012, I worked at the University of Oxford in collaboration with Julia Schnabel (Institute of Biomedical Engineering). Since January 2012, I continue this work at CNRS - Institut de Mathématiques de Toulouse. This work is also done in strong collaboration with François-Xavier Vialard (Université Dauphine, Paris). Applications are possible for any kind of images, in particular cerebral images, but also images of the lungs or the heart.Image registration consists in finding the optimal deformation to match source images onto target images with respect to spatial constrains. It has numerous applications in image analysis, from the comparison of shapes to the creation of atlases. Recent years have seen the development of diffeomorphic (smooth and invertible) registration techniques that allow large deformations. The formalism of the Large Deformation Diffeomorphic Metric Mapping (LDDMM - Beg et al., IJCV 2005) is one of the references in this context since it is designed to find geodesics between shapes, i.e., the shortest path between the shapes according to a metric. This formalism has therefore interesting statistical properties, in particular, for the creation of atlases. However, applications of the LDDMM framework on volumetric 3D medical images still remain limited for practical reasons. Interestingly, alternatives that are faster, requiring less memory or adapted to multi-modal images, have been proposed (e.g.: Avants et al., MIA 2008 / Vercauteren et al, MICCAI 2008 / ...) but none of them has been designed to explicitely estimate geodesic transformations between the registered images. In this context, I mainly work on the development of practical, but also mathematically justified, strategies for the geodesic registration of volumetric 3D cerebral images. I also work on the development of more general diffeomorphic registration strategies to estimate the motion of the lungs and the heart in CT/MR 3D images. A synthesis of my research activity in medical image registration between 2009 and 2018 can be found in my Habilitation thesis. 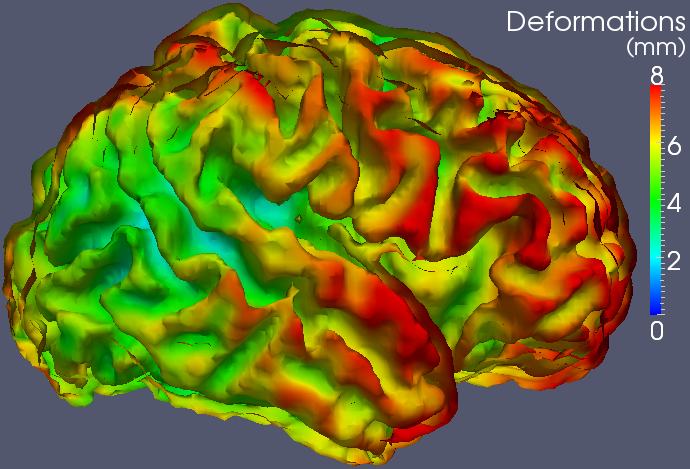  Software:The source code and executables of the multi-kernel LDDMM algorithm (Risser et al. TMI 2011) and beta executables of the geodesic shooting algorithm (Vialard et al. IJCV 2011) are available on sourceforge: sourceforge.net/projects/utilzreg/. Explanations about the software are given here.Some results on the cortical maturation:Registration of follow-up images acquired on pre-term babies at 33, 36 and 43 weeks of gestational age (first from 33 to 36 weeks and then from 36 to 43 weeks):2D slices 3D view - External face of the grey matter 3D view - Internal face of the grey matter Separation of the large and small scale contribution in the deformation estimated between 36 and 43 weeks: Estimated deformation -> the colors represent the total amount of deformations from 36 weeks. Estimated deformation -> the colors represent the large scale contribution in the deformations. Estimated deformation -> the colors represent the small scale contribution in the deformations. Other preliminary results:Registration of 2D binary images that highlights the potential of the LDDMMs (.avi file - OK using vlc).Registration of cardiac CT images (.avi file - OK using vlc). |


